Lab 2: Principal Component Analysis
This computer practical consists of three exercises; first we use factorextra package to visualise the results obtained by principal component analysis, and then we examine how the strength of existing redendancy in data affects the PCA results. In exercise 3, we will get a taste of principal component regression.
Exercise 1 (Iris data)
Here we use the same data (Iris data) illustrated in the Workshop, but will use the different function to perform PCA. First load the following package(s) into your workspace for visualisation
library(factoextra)
Task 1a
Use prcomp() to perfrom the PCA. For this run ?prcomp first. Check if the output of PCA agrees with the results obtained by running the command eigen() in the Workshop:PCA. Where do you get the eigenvalues, PC loadings and PC scores here?
Solution to Task 1
Click for solution
pr.out <- prcomp(iris[,-5] , scale =TRUE)
names(pr.out)
summary(pr.out)
# eigenvalues
pr.out$sdev^2
# PC loadings
pr.out$rotation
# PC score
pr.out$x
Task 1b
How do you interpret the Standard deviation of the principal components from the summary table? Can you manually compute the Proportion of Variance of the PC1 to PC4 by using the Standard deviation of the principal components?
Solution to Task 1b
Click for solution
The Standard deviation of the principal components is the square root of the corresponding eigenvalues.
sd_all <- summary(pr.out)$importance[1,]
sd_all^2/sum(sd_all^2)## PC1 PC2 PC3 PC4
## 0.729624454 0.228507618 0.036689219 0.005178709summary(pr.out)$importance[2,]## PC1 PC2 PC3 PC4
## 0.72962 0.22851 0.03669 0.00518
Type the following code to make a scree plot:
library(factoextra)
fviz_screeplot(pr.out, addlabels = TRUE)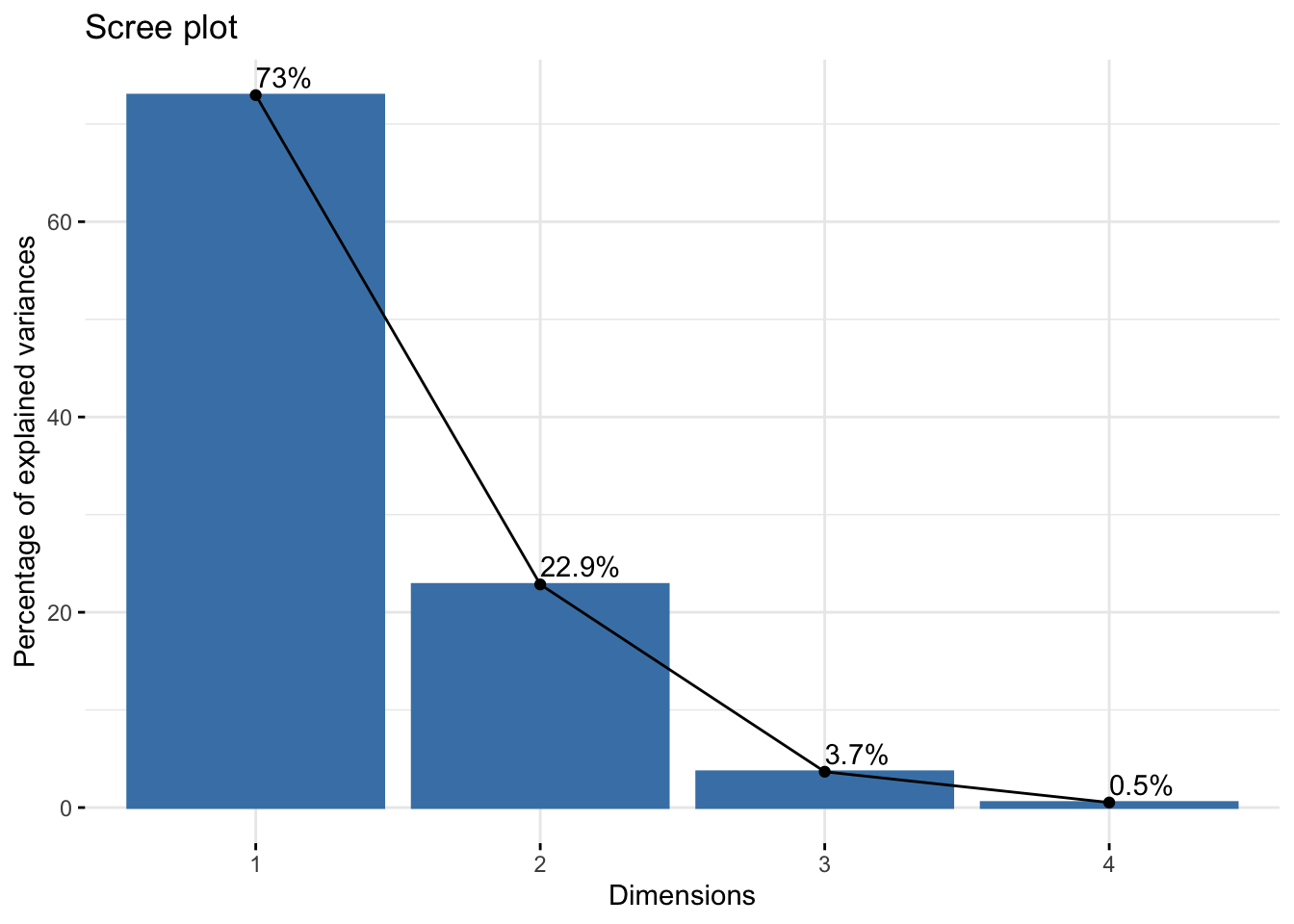
Correlation between variables and PCs, quality of representation (cos2) and contributions can be obtained respectively as
var <- get_pca_var(pr.out)
var$cor## Dim.1 Dim.2 Dim.3 Dim.4
## Sepal.Length 0.8901688 -0.36082989 0.27565767 0.03760602
## Sepal.Width -0.4601427 -0.88271627 -0.09361987 -0.01777631
## Petal.Length 0.9915552 -0.02341519 -0.05444699 -0.11534978
## Petal.Width 0.9649790 -0.06399985 -0.24298265 0.07535950var$cos2## Dim.1 Dim.2 Dim.3 Dim.4
## Sepal.Length 0.7924004 0.130198208 0.075987149 0.0014142127
## Sepal.Width 0.2117313 0.779188012 0.008764681 0.0003159971
## Petal.Length 0.9831817 0.000548271 0.002964475 0.0133055723
## Petal.Width 0.9311844 0.004095980 0.059040571 0.0056790544var$contrib## Dim.1 Dim.2 Dim.3 Dim.4
## Sepal.Length 27.150969 14.24440565 51.777574 6.827052
## Sepal.Width 7.254804 85.24748749 5.972245 1.525463
## Petal.Length 33.687936 0.05998389 2.019990 64.232089
## Petal.Width 31.906291 0.44812296 40.230191 27.415396Run the code above and check to see that these values are in agreement with the results in the step-by-step approach in the tutorials.
You can plot the contribution of the variables to PC1 as follows:
fviz_contrib(pr.out, choice = "var", axes = 1, top = 10)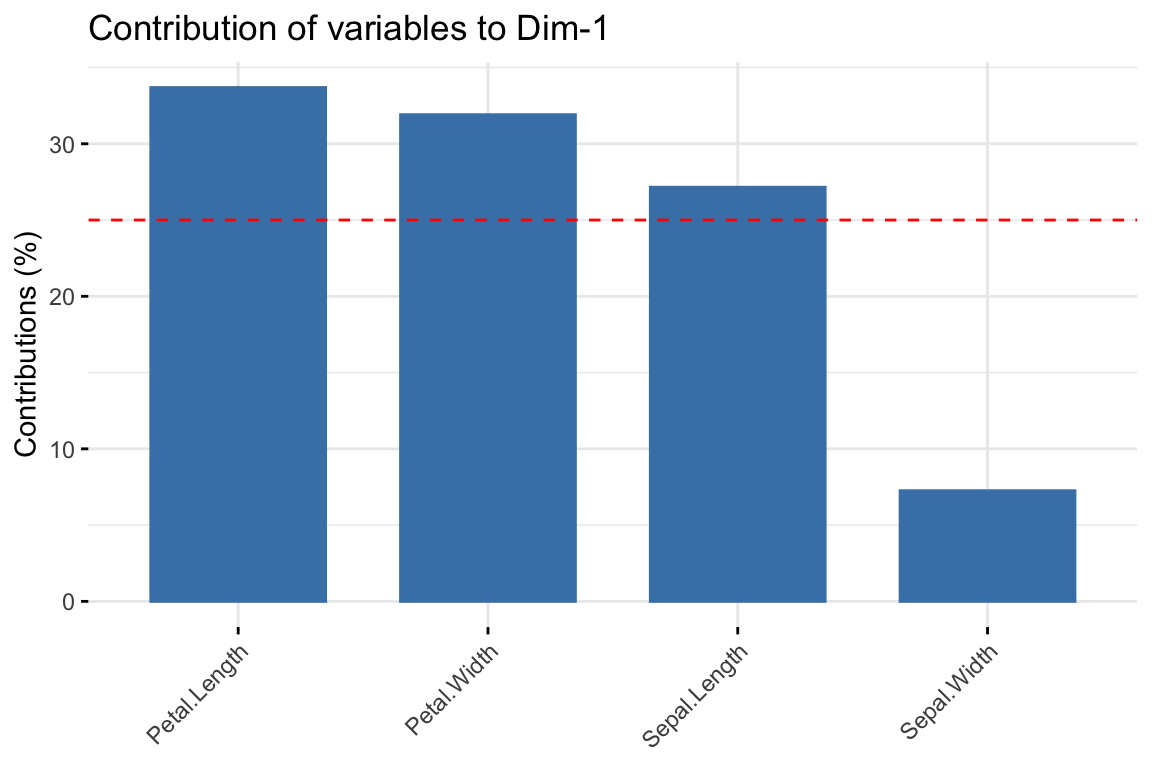
Task 1c
Modify the code to plot the contribution of the variables to PC2.
Click for solution
# Contributions of variables to PC2
fviz_contrib(pr.out, choice = "var", axes = 2, top = 10)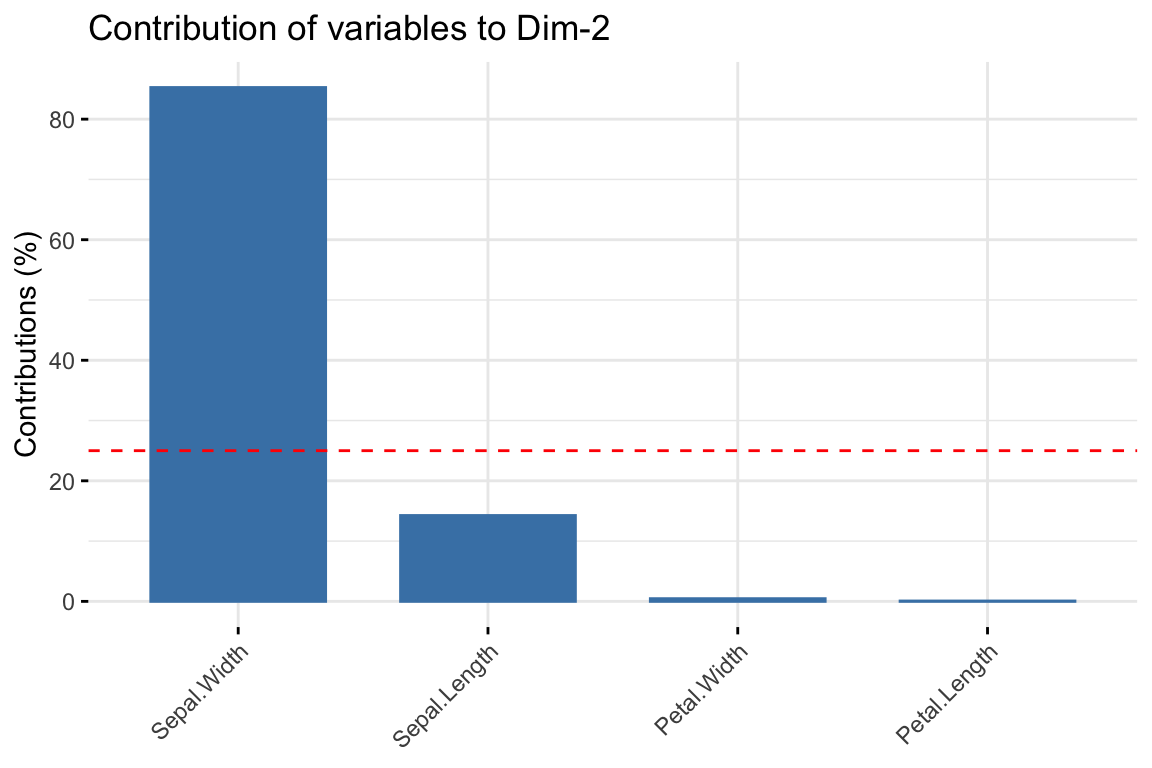
There are various ways to make a biplot. Try the following codes:
biplot (pr.out , scale =0)
abline(h=0, col="gray",lty=3, lwd =2)
abline(v=0, col="gray",lty=3, lwd =2)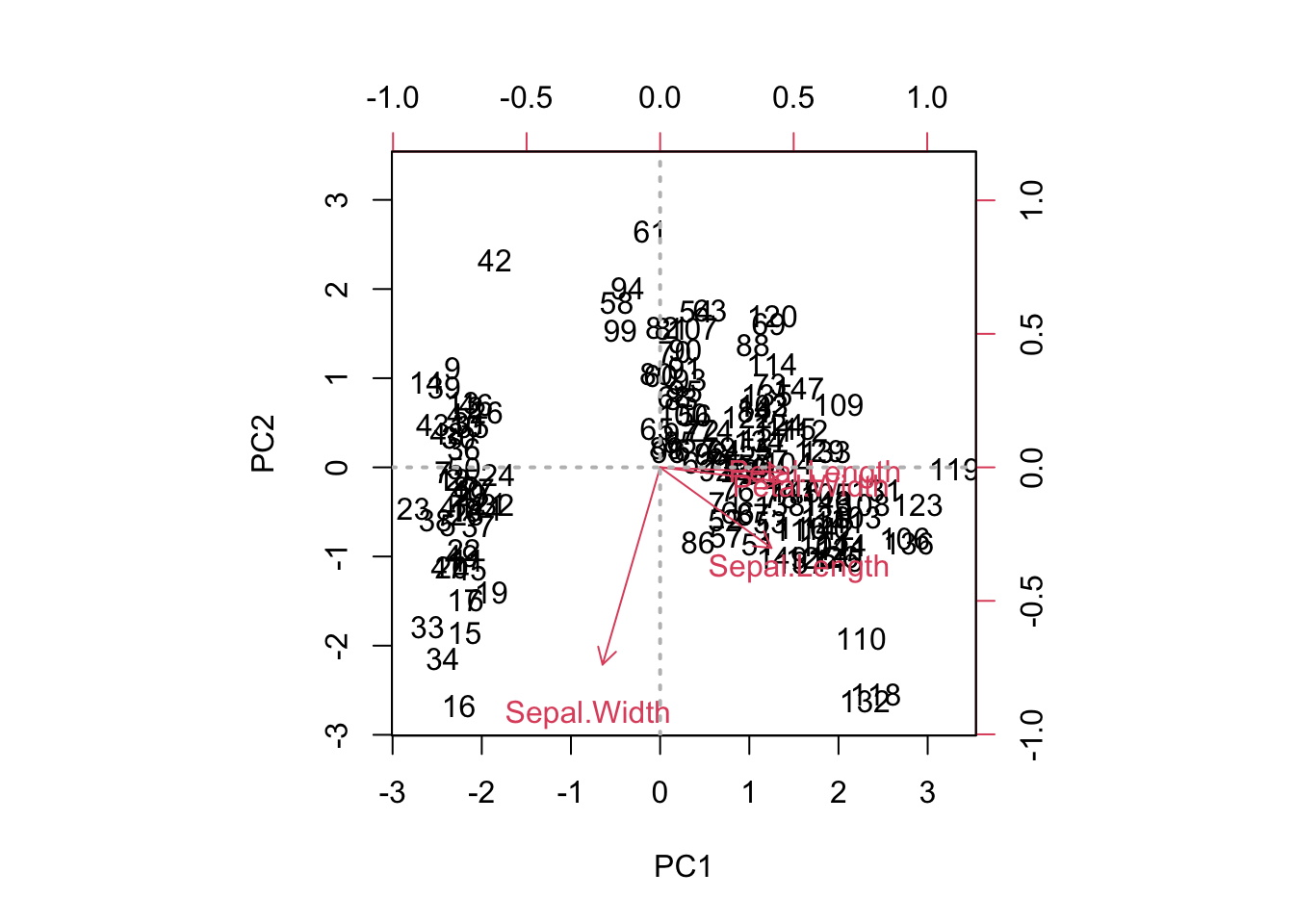
and a more elegant one.
fviz_pca_biplot(pr.out,
col.ind = iris$Species, palette = "jco",
addEllipses = TRUE, label = "var",
col.var = "black", repel = TRUE,
legend.title = "Species")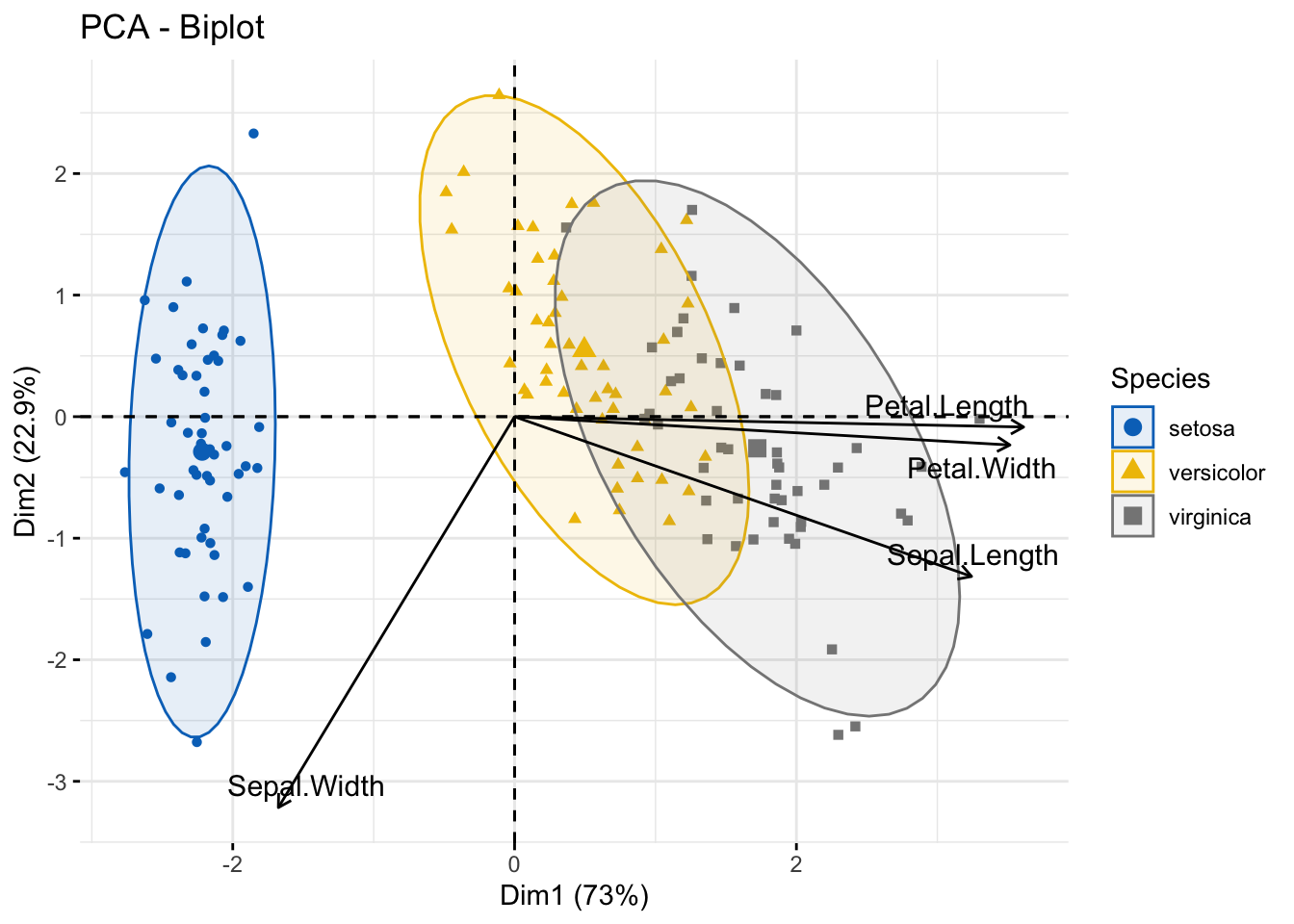 You can obtain alternative visualisation of a biplot using
You can obtain alternative visualisation of a biplot using autoplot function in the ggfortifyCheck package (please check these out at your own convenience).
Exercise 2
Generate n = 200 observations and p=30 variables from a multivariate normal distribution with correlation matrix given as \(corr(x_{i},x_{j}) = \rho^{|i-j|}\). You can create the covariance matrix using
covmat <- function(rho, p) {
rho^(abs(outer(seq(p), seq(p), "-")))
}and use the mvrnorm function in MASS package to generate the multivariate Gaussian data.
Task 2a
Take \(\rho=0.95\) and carry out PCA. For reproducibility, take the pseudo-number generator, seed =123. How many PCs is sufficient for this data? Comment on your general observation.
(Hint: Use the following codes to generate the data called dd.
library(MASS)
set.seed(123)
p = 30
n = 200
cov.e = covmat(0.95, p)
mean.e = rep(0, p)
dd <- mvrnorm(n, mean.e, cov.e)
Click for solution
prr <- prcomp(dd)
summary(prr)## Importance of components:
## PC1 PC2 PC3 PC4 PC5 PC6 PC7
## Standard deviation 4.1415 2.2933 1.3464 1.00341 0.77935 0.63838 0.48520
## Proportion of Variance 0.6179 0.1895 0.0653 0.03627 0.02188 0.01468 0.00848
## Cumulative Proportion 0.6179 0.8073 0.8726 0.90886 0.93074 0.94542 0.95390
## PC8 PC9 PC10 PC11 PC12 PC13 PC14
## Standard deviation 0.41615 0.40216 0.37139 0.29564 0.29060 0.26536 0.2469
## Proportion of Variance 0.00624 0.00583 0.00497 0.00315 0.00304 0.00254 0.0022
## Cumulative Proportion 0.96014 0.96597 0.97093 0.97408 0.97712 0.97966 0.9819
## PC15 PC16 PC17 PC18 PC19 PC20 PC21
## Standard deviation 0.24380 0.22571 0.21583 0.20360 0.18959 0.18087 0.17804
## Proportion of Variance 0.00214 0.00184 0.00168 0.00149 0.00129 0.00118 0.00114
## Cumulative Proportion 0.98400 0.98583 0.98751 0.98900 0.99030 0.99148 0.99262
## PC22 PC23 PC24 PC25 PC26 PC27 PC28
## Standard deviation 0.1744 0.16711 0.16001 0.15574 0.15240 0.14561 0.1390
## Proportion of Variance 0.0011 0.00101 0.00092 0.00087 0.00084 0.00076 0.0007
## Cumulative Proportion 0.9937 0.99472 0.99564 0.99652 0.99735 0.99812 0.9988
## PC29 PC30
## Standard deviation 0.12848 0.12825
## Proportion of Variance 0.00059 0.00059
## Cumulative Proportion 0.99941 1.00000plot(summary(prr)$importance[2,], type="b", xlab="PCs", ylab="Variability explained")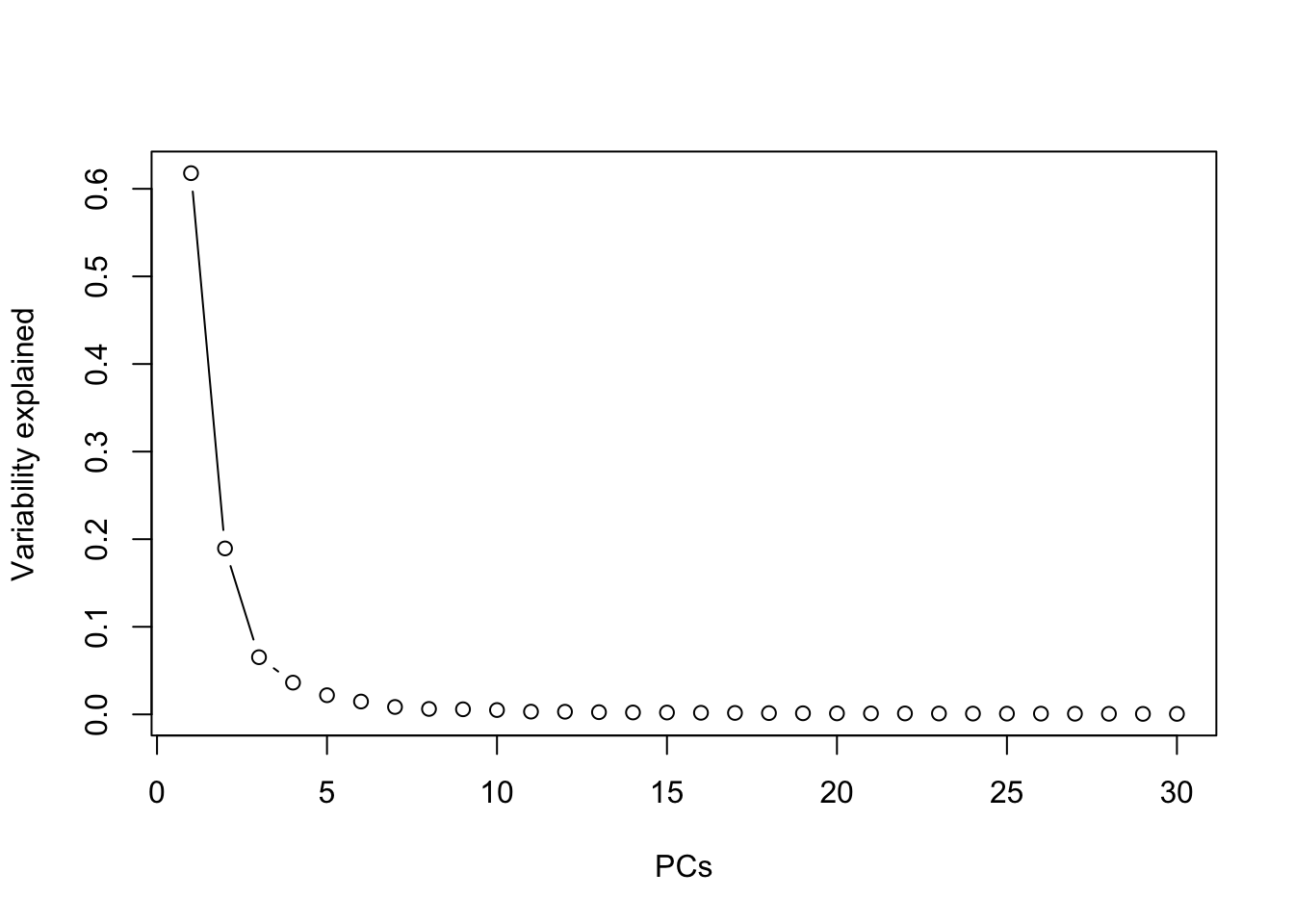
Task 2b
Take \(\rho=0.2\) and carry out PCA. As done before, use the pseudo-number generator, seed = 123 for reproducibility. How many PCs are sufficient for this data? Compare your results with the case of \(\rho=0.95\).
Click for solution
library(MASS)
set.seed(123)
p = 30
n = 200
cov.e = covmat(0.2, p)
mean.e = rep(0, p)
dd <- mvrnorm(n, mean.e, cov.e)
prr <- prcomp(dd)
summary(prr)## Importance of components:
## PC1 PC2 PC3 PC4 PC5 PC6 PC7
## Standard deviation 1.46267 1.33775 1.32938 1.28976 1.22917 1.17781 1.17183
## Proportion of Variance 0.07223 0.06042 0.05966 0.05616 0.05101 0.04683 0.04636
## Cumulative Proportion 0.07223 0.13265 0.19231 0.24847 0.29948 0.34631 0.39267
## PC8 PC9 PC10 PC11 PC12 PC13 PC14
## Standard deviation 1.16283 1.13430 1.1074 1.05365 1.03779 1.01332 0.99861
## Proportion of Variance 0.04565 0.04344 0.0414 0.03748 0.03636 0.03467 0.03367
## Cumulative Proportion 0.43832 0.48176 0.5232 0.56065 0.59701 0.63167 0.66534
## PC15 PC16 PC17 PC18 PC19 PC20 PC21
## Standard deviation 0.96032 0.92220 0.91021 0.88108 0.85584 0.83619 0.81532
## Proportion of Variance 0.03114 0.02871 0.02797 0.02621 0.02473 0.02361 0.02244
## Cumulative Proportion 0.69648 0.72519 0.75316 0.77937 0.80410 0.82770 0.85015
## PC22 PC23 PC24 PC25 PC26 PC27 PC28
## Standard deviation 0.78841 0.75942 0.74078 0.71840 0.70589 0.69243 0.66375
## Proportion of Variance 0.02099 0.01947 0.01853 0.01742 0.01682 0.01619 0.01487
## Cumulative Proportion 0.87113 0.89060 0.90913 0.92655 0.94338 0.95956 0.97444
## PC29 PC30
## Standard deviation 0.62502 0.60544
## Proportion of Variance 0.01319 0.01238
## Cumulative Proportion 0.98762 1.00000plot(summary(prr)$importance[2,], type="b", xlab="PCs", ylab="Variability explained")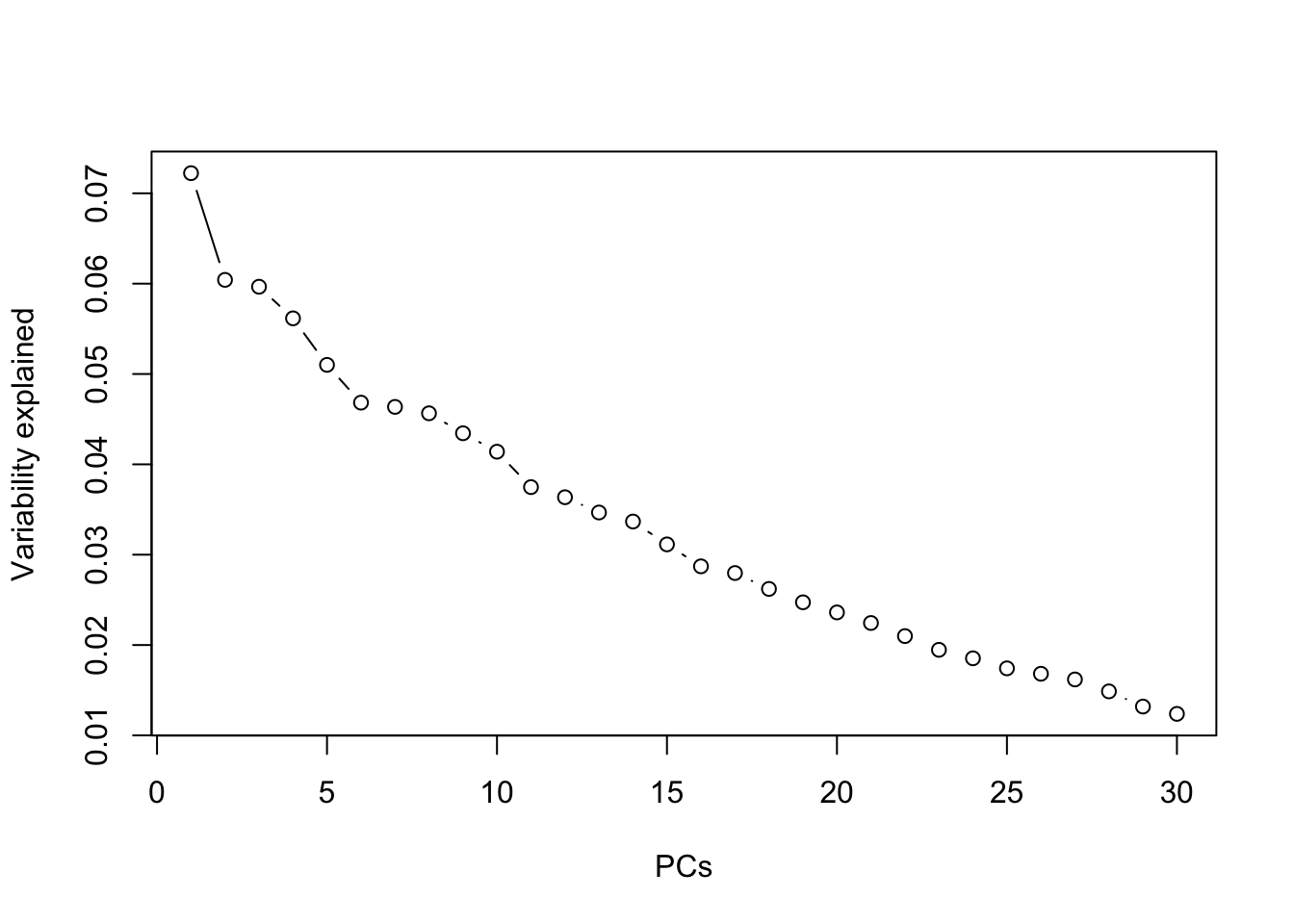
The summary shows that 19 components are enough to keep 80% of the variability in data. This shows that the higher the correlation, the higher the redundancy in the data in which case PCA becomes more useful and gives more clear interpretation.
Exercise 3
One of the many uses of PCA is principal component regression (PCR). We have seen that Sepal length is highly correlated with Petal length and Petal width.
Task 3a
Suppose that we want to carryout regression analysis between Sepal length (response variable) and the three remaining variables (covariates). Then we can use PCA to reduce the correlation. Perform the PCA and find the principal scores.
Click for solution
iris.pca <- prcomp(iris[,2:4], scale=TRUE)
iris.pca$x## PC1 PC2 PC3
## [1,] -2.1248387 -0.139743260 0.008370721
## [2,] -1.6451988 0.900407772 -0.054584971
## [3,] -1.8737775 0.498821958 -0.070033694
## [4,] -1.7044039 0.677902966 -0.001362832
## [5,] -2.2207666 -0.347773466 0.020961859
## [6,] -2.2314161 -1.103311779 -0.001635822
## [7,] -1.9454276 0.024274998 -0.095352466
## [8,] -1.9921879 0.053812348 0.036410583
## [9,] -1.5492708 1.108437978 -0.067176109
## [10,] -1.7878870 0.721914915 0.089769216
## [11,] -2.2799718 -0.570278271 0.074183997
## [12,] -1.9554651 0.039337749 0.077041583
## [13,] -1.7286818 0.944419721 0.036547077
## [14,] -1.8388502 0.987843518 -0.085345923
## [15,] -2.6779242 -1.150945093 -0.009935588
## [16,] -2.7845016 -2.114513613 -0.019942131
## [17,] -2.3783073 -1.045413383 -0.164159822
## [18,] -2.0413556 -0.183755209 -0.082761327
## [19,] -2.2189712 -0.851269624 0.076905088
## [20,] -2.2924168 -0.822320426 -0.004356912
## [21,] -1.9187423 0.024863150 0.117672583
## [22,] -2.1130058 -0.658302169 -0.108080099
## [23,] -2.3676579 -0.289875070 -0.141562142
## [24,] -1.5723652 0.100857509 -0.168314699
## [25,] -1.8452967 -0.004086048 0.198934583
## [26,] -1.5717531 0.871458574 0.026677030
## [27,] -1.7884990 -0.048686149 -0.105222513
## [28,] -2.0881159 -0.154217859 0.049001721
## [29,] -2.0289107 0.068286947 -0.004220418
## [30,] -1.7636091 0.455398161 0.051859306
## [31,] -1.6676811 0.663428368 0.039268168
## [32,] -1.8252218 -0.034211550 -0.145853513
## [33,] -2.7471668 -1.358387147 0.215680599
## [34,] -2.7963345 -1.595954703 0.096508689
## [35,] -1.7044039 0.677902966 -0.001362832
## [36,] -1.9105003 0.513296557 -0.110664695
## [37,] -2.1615615 -0.125268661 -0.032260280
## [38,] -2.3042497 -0.303761517 0.112093907
## [39,] -1.6819216 0.914882371 -0.095215971
## [40,] -1.9921879 0.053812348 0.036410583
## [41,] -2.0780784 -0.169280610 -0.123392328
## [42,] -0.9269427 2.327081865 -0.274485987
## [43,] -1.8737775 0.498821958 -0.070033694
## [44,] -1.7174609 -0.344740253 -0.274895471
## [45,] -2.0620425 -0.924230771 0.067035040
## [46,] -1.5617157 0.856395823 -0.145717019
## [47,] -2.3391770 -0.792783076 0.127406136
## [48,] -1.8370547 0.484347359 -0.029402694
## [49,] -2.2799718 -0.570278271 0.074183997
## [50,] -1.9329827 0.276317153 -0.016811556
## [51,] 0.3765942 -0.521457794 0.217835736
## [52,] 0.3866316 -0.536520545 0.045441688
## [53,] 0.6294508 -0.386388735 0.195374550
## [54,] 0.8994033 1.496148204 -0.088769462
## [55,] 0.8070663 0.281125681 0.035708135
## [56,] 0.6033775 0.383624178 0.177341231
## [57,] 0.4476323 -0.817511898 0.048162778
## [58,] 0.2959666 1.521476037 -0.087199181
## [59,] 0.5441723 0.161119373 0.230563369
## [60,] 0.5624516 0.634490029 -0.170167957
## [61,] 0.7531242 2.324647664 -0.056301734
## [62,] 0.4683191 -0.077036336 -0.101633589
## [63,] 0.7448822 1.836214257 0.172035544
## [64,] 0.6643781 0.102632825 0.180062321
## [65,] 0.1769442 0.305865362 -0.175746633
## [66,] 0.3623537 -0.270003791 0.083351597
## [67,] 0.5784876 -0.120460133 0.020259411
## [68,] 0.3019651 0.781588627 0.275622236
## [69,] 1.3459114 1.543781517 -0.080469695
## [70,] 0.5038585 1.182586288 0.078045911
## [71,] 0.7472491 -0.711980189 -0.106061456
## [72,] 0.4197634 0.455997173 -0.025813770
## [73,] 1.2050187 0.861792503 0.119827721
## [74,] 0.5933400 0.398686929 0.349735279
## [75,] 0.4340039 0.204543170 0.108670369
## [76,] 0.4582817 -0.061973585 0.070760459
## [77,] 0.7970289 0.296188432 0.208102183
## [78,] 0.9290676 -0.280857025 0.041150316
## [79,] 0.6744155 0.087570074 0.007668273
## [80,] 0.1775563 1.076466427 0.019245096
## [81,] 0.5630637 1.405091093 0.024823772
## [82,] 0.4428579 1.463577641 0.075324820
## [83,] 0.3954856 0.722513927 0.012096139
## [84,] 1.1700913 0.372770943 0.135139949
## [85,] 0.5784876 -0.120460133 0.020259411
## [86,] 0.2782587 -0.996592907 -0.020508084
## [87,] 0.5560052 -0.357439537 0.114112550
## [88,] 1.0462946 1.438249808 0.073754539
## [89,] 0.2646303 0.025462161 0.039999506
## [90,] 0.7075474 1.080087791 -0.063587185
## [91,] 0.6750276 0.858171138 0.202660002
## [92,] 0.5317273 -0.090922783 0.152022459
## [93,] 0.5281364 0.916069534 0.040136001
## [94,] 0.3918946 1.729506244 -0.099790319
## [95,] 0.5891370 0.635078181 0.042857092
## [96,] 0.2178701 0.054999511 0.171762555
## [97,] 0.3972811 0.219017768 0.068039368
## [98,] 0.4340039 0.204543170 0.108670369
## [99,] 0.1733533 1.312857679 -0.287633091
## [100,] 0.4564862 0.441522574 0.014817230
## [101,] 1.6763760 -1.401789225 -0.243822651
## [102,] 1.4205404 0.240735097 -0.138256195
## [103,] 1.5935050 -0.587176212 0.042301126
## [104,] 1.3288155 -0.203686362 0.181213131
## [105,] 1.6402652 -0.616713562 -0.089461922
## [106,] 1.8505646 -0.688498405 0.326718127
## [107,] 1.2250935 0.831667001 -0.224960376
## [108,] 1.5858751 -0.305008555 0.465630132
## [109,] 1.7859730 0.599485265 0.212110578
## [110,] 1.4253148 -2.040354443 -0.165418236
## [111,] 1.0243836 -0.843427884 -0.166432551
## [112,] 1.4939861 0.211785899 -0.056994194
## [113,] 1.4466138 -0.529277816 -0.120222875
## [114,] 1.6591566 0.627258159 -0.295201519
## [115,] 1.7420276 -0.187354854 -0.581325296
## [116,] 1.3482783 -1.004412928 -0.358566695
## [117,] 1.1961647 -0.397241969 0.153173269
## [118,] 1.2033466 -2.411226602 0.376946186
## [119,] 2.5114110 0.012174726 0.215982479
## [120,] 1.5295254 1.471408523 0.122685306
## [121,] 1.4951695 -1.062311324 -0.196042694
## [122,] 1.3346499 0.017642139 -0.298059105
## [123,] 1.9956603 -0.242900642 0.433298899
## [124,] 1.2636118 0.313696243 -0.128386147
## [125,] 1.2322754 -1.182317633 -0.001187460
## [126,] 1.1879227 -0.885675376 0.381510547
## [127,] 1.1309610 0.120140636 -0.156426009
## [128,] 0.9758279 -0.310394375 -0.090612732
## [129,] 1.6751925 -0.127692002 -0.104774151
## [130,] 1.1393670 -0.352641868 0.457330366
## [131,] 1.6918405 -0.112041099 0.280644946
## [132,] 0.9262121 -2.279778908 0.437317281
## [133,] 1.7586756 -0.171703951 -0.195906199
## [134,] 0.9906803 0.208752686 0.238863136
## [135,] 1.2826673 0.596452053 0.507967908
## [136,] 1.8339167 -0.704149308 -0.058700970
## [137,] 1.3500738 -1.507909087 -0.302623466
## [138,] 1.1002367 -0.605272175 0.165764407
## [139,] 0.9391051 -0.295919776 -0.131243732
## [140,] 1.3139630 -0.722833423 -0.148262737
## [141,] 1.6378577 -0.883818468 -0.340396881
## [142,] 1.3707607 -0.767433524 -0.452419834
## [143,] 1.4205404 0.240735097 -0.138256195
## [144,] 1.5686151 -1.091260522 -0.114780694
## [145,] 1.5662076 -1.358365428 -0.365715652
## [146,] 1.5034114 -0.573877917 -0.424379972
## [147,] 1.5756736 0.671270108 -0.204069471
## [148,] 1.2529623 -0.441842070 -0.150983828
## [149,] 1.1931451 -1.434947940 -0.292753418
## [150,] 1.0492735 -0.339343573 -0.009350732
Task 3b
Run a linear regression by using lm() command to carryout the principal component regression (PCR, not PCA!). Use just the first 2 principal components as covariates in the linear regression model. As explained before, the response variable should be Sepal length.
Click for solution
Z = iris.pca$x[,1:2] # select the first two PCs
iris.lm <- lm(iris$Sepal.Length~Z)
iris.lm##
## Call:
## lm(formula = iris$Sepal.Length ~ Z)
##
## Coefficients:
## (Intercept) ZPC1 ZPC2
## 5.8433 0.4230 -0.4348
Now we transform this coefficient vector to the scale of the original covariates as follows. This gives the final PCR estimator whose dimension is equal to the total number of original covariates.
iris.pca$rotation[,1:2]%*%matrix(coef(iris.lm)[-1], ncol=1)## [,1]
## Sepal.Width 0.2173767
## Petal.Length 0.3853085
## Petal.Width 0.4150270
Task 3c
In practice, we do not need to do PCR ourselves in this way, as it has been implemented in the pls R package. Carryout PCR using the pcr function in R (check HERE) and compare the results.
Click for solution
library(pls)
iris.pcr <- pcr(Sepal.Length~ Sepal.Width+Petal.Length+Petal.Width, 2,
scale=TRUE, data=iris) # 2 stands for number of components,
coef(iris.pcr)## , , 2 comps
##
## Sepal.Length
## Sepal.Width 0.2173767
## Petal.Length 0.3853085
## Petal.Width 0.4150270